Early detection of Ganoderma
A survey indicated that BSR was present in more than 59.6% (632 out of 1046 estates surveyed in 2010) of the oil palm estates in Malaysia.
Unfortunately, many planters especially smallholders did not realise that their oil palms were infected with Ganoderma, even in situations where the disease was obvious.
Several methods for early detection of Ganoderma in oil palm have been developed, as follows:
- Culture-based, e.g. Drilling and Ganoderma Selective Medium (GSM)
- Molecular method, e.g. PCR
- Bio-sensor, e.g. GanoSken tomography
Drilling and GSM method
GSM can be used to confirm the presence of Ganoderma disease in oil palm. With GSM, it is possible to detect infected oil palm without any obvious symptom of infection externally (asymptomatic palms) using a drilling technique. Studies indicate that 10-16% of seemingly healthy oil palm are already infected with Ganoderma fungus.

The suspected palm is drilled at 30 cm depth at the palm base using a drill bit (11 mm diameter, 45 cm long) that is attached to an engine drill
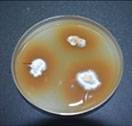
Stem tissues are placed onto the GSM, incubated for 5-8 days. The white mycelium that grows from the tissue is then transferred onto potato dextrose agar (PDA) plate

Petri plate culture of Ganoderma. Identification is based on the morphological characteristics using microscope
GanoSkenTM tomography method
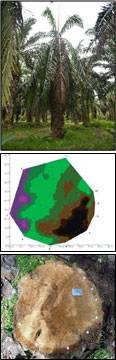
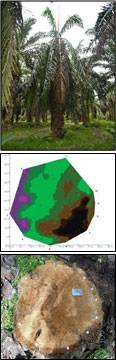
Tomography refers to the cross sectional imaging of an object from either transmission or reflection data collected by exposing the object under study to the wave source from various directions. The tomography technology is a non-invasive tool designed for assessing a tree’s decay and degradation. The equipment consists of Sound Sensor and Tomography software called GanoSkenTM. Sound sensors are installed around a circumference of the oil palm stem under study. Sound wave is emitted on one sensor and time of flight of the sound propagation from the emitter to other sensors is calculated. These sound lines are then used to construct the tomography image of the stem. The detection together with the location and the size of the decay and degradation will allow the expert on deciding effective disease treatments on the oil palm trunk.
A field evaluation was carried out on 30 cross sections of oil palm stems that were classified into two groups: healthy-looking palms (H) and diseased (infected) palms (I). GanoSkenTM tomography images were produced and the size and site (location) of Ganoderma infection in oil palm stem are confirmed. The images of real cross sections correlated with tomography images. Dark brown areas indicate the intact and healthy stem, while purple and greenish areas indicate Ganoderma infection. Ganoderma infection on the oil palm stems were confirmed using the GSM.
Results showed that GanoSkenTM tomography is capable in detecting and identifying Ganoderma infection in healthy-looking (asymptomatic) oil palm. The GanoSkenTM tomography is capable of performing an early detection of Ganoderma infection in oil palm trunk especially in healthy-looking oil palms.

Detection of Ganoderma using GanoskenTM. GanoskenTM tomography images were produced
Molecular PCR-DNA method
The use of Polymerase Chain Reaction (PCR) technique for detection offers early detection and screening of pathogenic Ganoderma in oil palm.
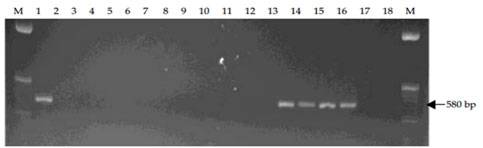
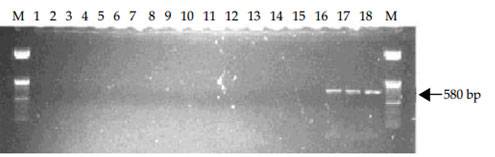
MPOB has develop a forward primer, PER44-123 paired with a reverse primer LR1. The primers amplify partial ITS region of the ribosomal RNA and produce 580 bp amplified fragments. These primers are specific to pathogenic Ganoderma and only detect G. boninense, G. zonatum and G. miniatocinctum and shown not to detect other Ganoderma species such as G. tornatum, G. philippii, G. lucidum, G. weberianum, G. applanatum, G. oregonense, G. chalceum, G. pfeifferi, and G. resinaceum (Figure A) and other several other fungal/oomycetes species including fungus found on rotten palm parts (roots, stem and fruit bunches) and soil such as Aspergillus, Botryodiplodia, Gymnopilus, Fusarium, Marasmius, Pythium, Penicillium, Pycnoporus, Rhizopus, Rhizoctonia, Schizophyllum, Thielaviopsis, Trichoderma and Volvariella (Figure B).
Sensitivity of the primers for PCR was high, and DNA was detectable at concentrations as low as 10 pg(µl)-1.
Figure A. Specificity tests of PER44-123 and LR1 with DNA extracted from G. boninense and 11 other species of Ganoderma. Lane 1, G. boninense; Lane 2, G. philippii; Lanes 3-4, G. lucidum; Lane 5, G. weberianum; Lanes 6-7, G. applanatum; Lane 8, G. oregonense; Lane 9, G. chalceum; Lane 10, G. pfeifferi; Lanes 11-12, G. tornatum; Lanes 13-14, G. miniatocinctum; Lanes 15-16, G. zonatum; Lane 17, G. resinaceum and Lane 18, negative control of sterile distilled water. M is 50 bp DNA ladder.
Figure B. Specificity tests of primers PER44-123 and LR1 with DNA extracted from three species of Ganoderma and 14 fungi/oomycete. Lane 1, negative control of sterile distilled water; Lanes 2-18, Aspergillus sp., Botryodiplodia theobromae, Gymnopilus sp., Fusarium sp., Marasmius palmivorus, Pythium sp., Penicillium sp., Pycnoporus sp., Rhizopus sp., Rhizoctonia sp., Schizophyllum commune, Thielaviopsis paradoxa, Trichoderma sp., Volvariella sp., G. miniatocinctum, G. zonatum and G. boninense. M is 50 bp DNA ladder.